4.4 Are these SNPs in LD?
If we run table on each SNP column, we can see which alleles exist at SNP1 and SNP2.
- SNP1 can be
AorG - SNP2 can be
CorT
##
## A G
## 3456 1552##
## C T
## 2825 2183If these two SNPs were in perfect LD, we’d expect to see only two haplotypes in our data (Fig. 4A).
AC: If someone carries anAat SNP1, they will always carry aCat SNP2.GT: If they carry aGat SNP1, they will always carry aTat SNP2.
If these two SNPs were in linkage equilibrium, the allele at SNP1 would give us no information about SNP2. We would expect to see all four possible haplotypes, in amounts proportional to the component allele frequencies (Fig. 4B).
ACATGCGT
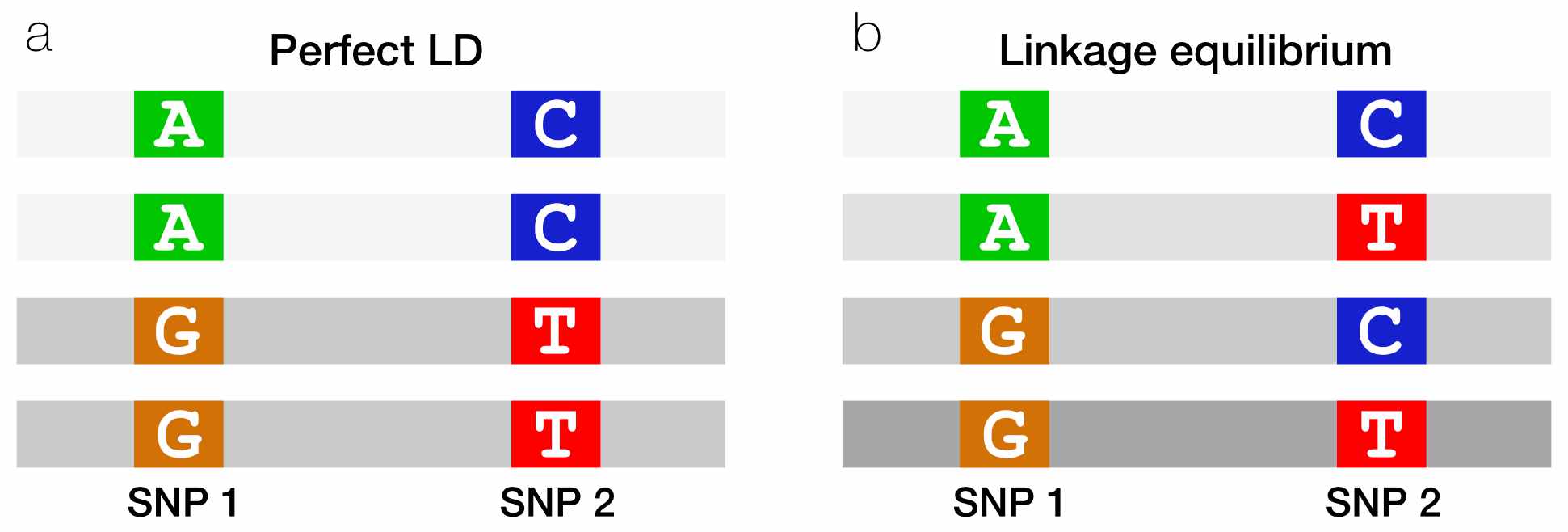
Fig. 4. When two SNPs are in perfect LD, seeing an allele on one haplotype perfectly predicts which allele is on the other haplotype.