9.19 Plotting PBS trees
Another useful way to visualize PBS is by comparing trees for the top PBS outliers to the genome-wide average tree. Run the code blocks below to plot these trees:
# create average tree
tr_mean <- rtree(n = 3,
rooted = FALSE,
br = c(mean(pbs$T_rps_png),
mean(pbs$T_rps_chb),
mean(pbs$T_png_chb)))
# plot average tree
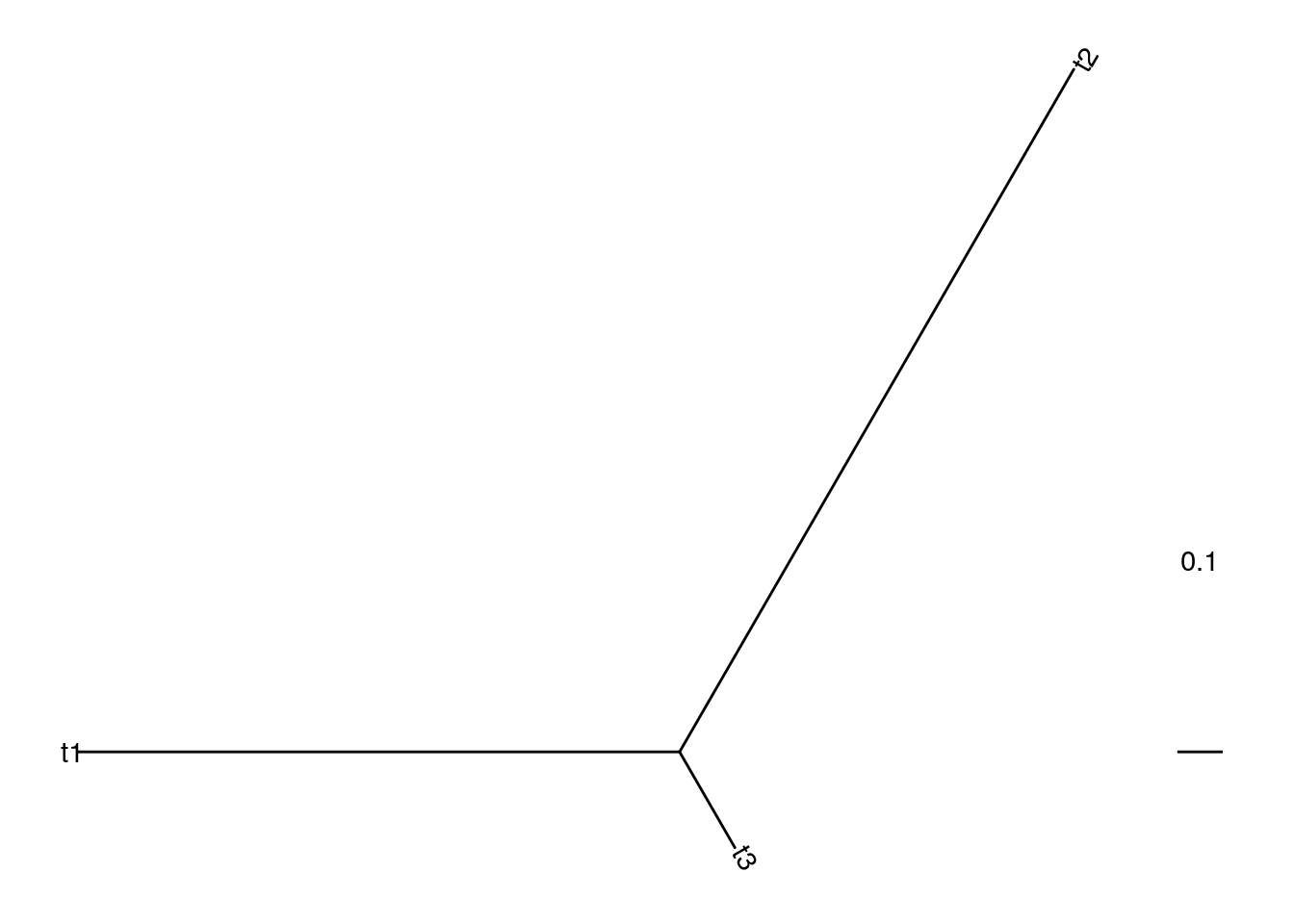
ggtree(tr_mean, layout = "daylight") +
geom_treescale(width = 0.1) +
geom_tiplab(label = c("RPS", "PNG", "CHB"))## Average angle change [1] 0.333333333333333## Average angle change [2] 2.22044604925031e-16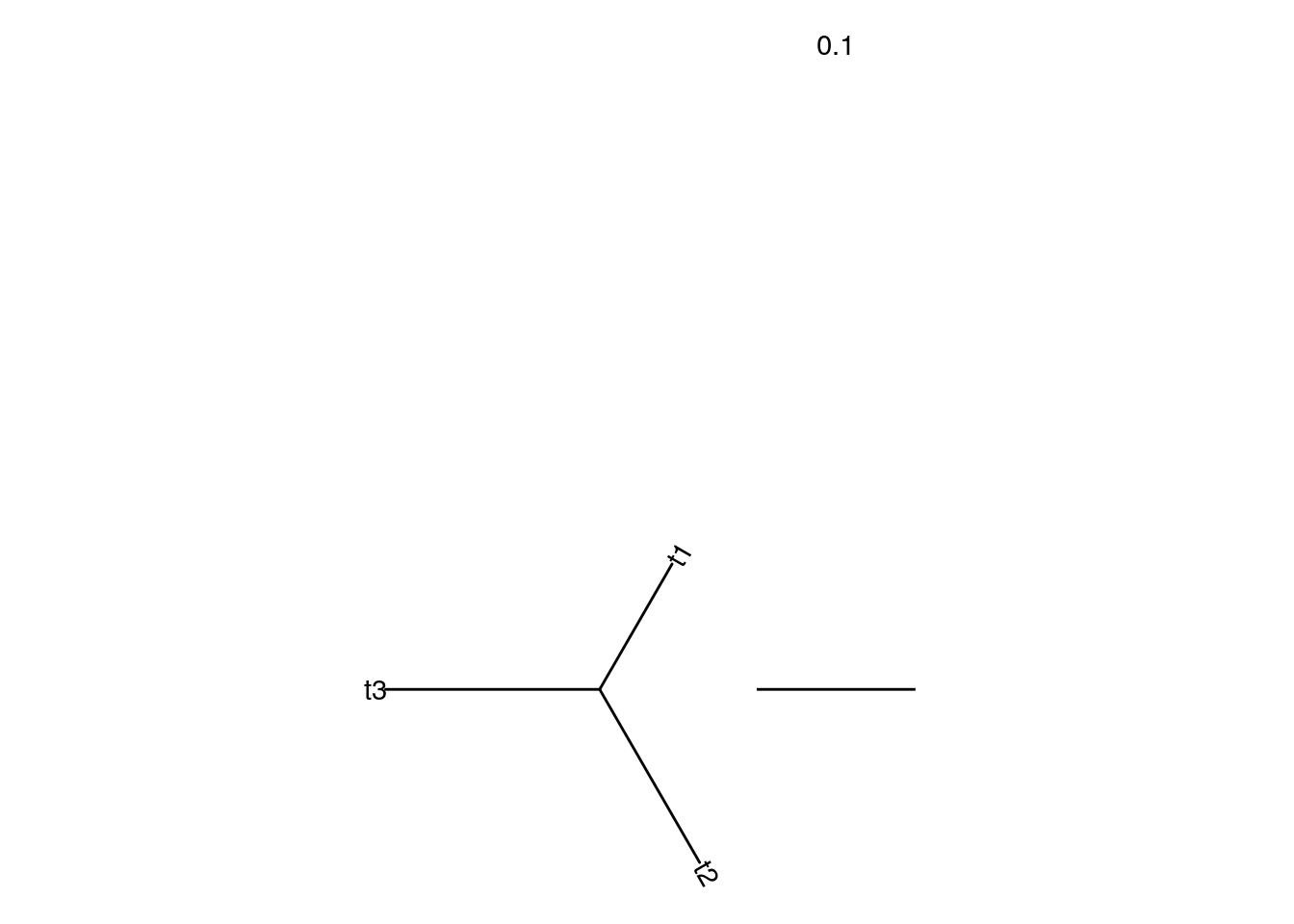
# create tree for top snp
tr_top <- rtree(n = 3,
rooted = FALSE,
br = c(pbs[1,]$T_rps_png,
pbs[1,]$T_rps_chb,
pbs[1,]$T_png_chb))
# plot top snp tree
ggtree(tr_top, layout = "daylight") +
geom_treescale(width = 0.1) +
geom_tiplab(label = c("RPS", "PNG", "CHB"))## Average angle change [1] 0.333333333333333## Average angle change [2] 2.22044604925031e-16