4.3 Setup
We’ll measure LD between two SNPs called in the 1000 Genomes Project dataset:
- rs28574812 (
chr21:15012619) - rs2251399 (
chr21:15013185)
We’ve preprocessed the original 1000 Genomes data such that every line in the table below represents one haplotype in the 1000 Genomes database. Load the pre-processed data by running the code below.
# read data
haplotypes <- read.table("snp_haplotypes.txt", header = TRUE)
# preview data
head(haplotypes)## sample haplotype snp1_allele snp2_allele
## 1 HG00096 hap_1 A C
## 2 HG00096 hap_2 A C
## 3 HG00097 hap_1 A C
## 4 HG00097 hap_2 A C
## 5 HG00099 hap_1 A C
## 6 HG00099 hap_2 A CThe columns in this table are:
sample: Name of the individual who was sequencedhaplotype: Haplotype (i.e., the maternal or paternal chromosome) that the SNP is onsnp1_allele: Genotype at SNP1 on this haplotypesnp2_allele: Genotype at SNP2 on this haplotype
Note that there are 2,504 samples in the 1000 Genomes Project but 5,008 total lines in the table. This is because there are two lines per individual – one for each of their maternal and paternal haplotypes.
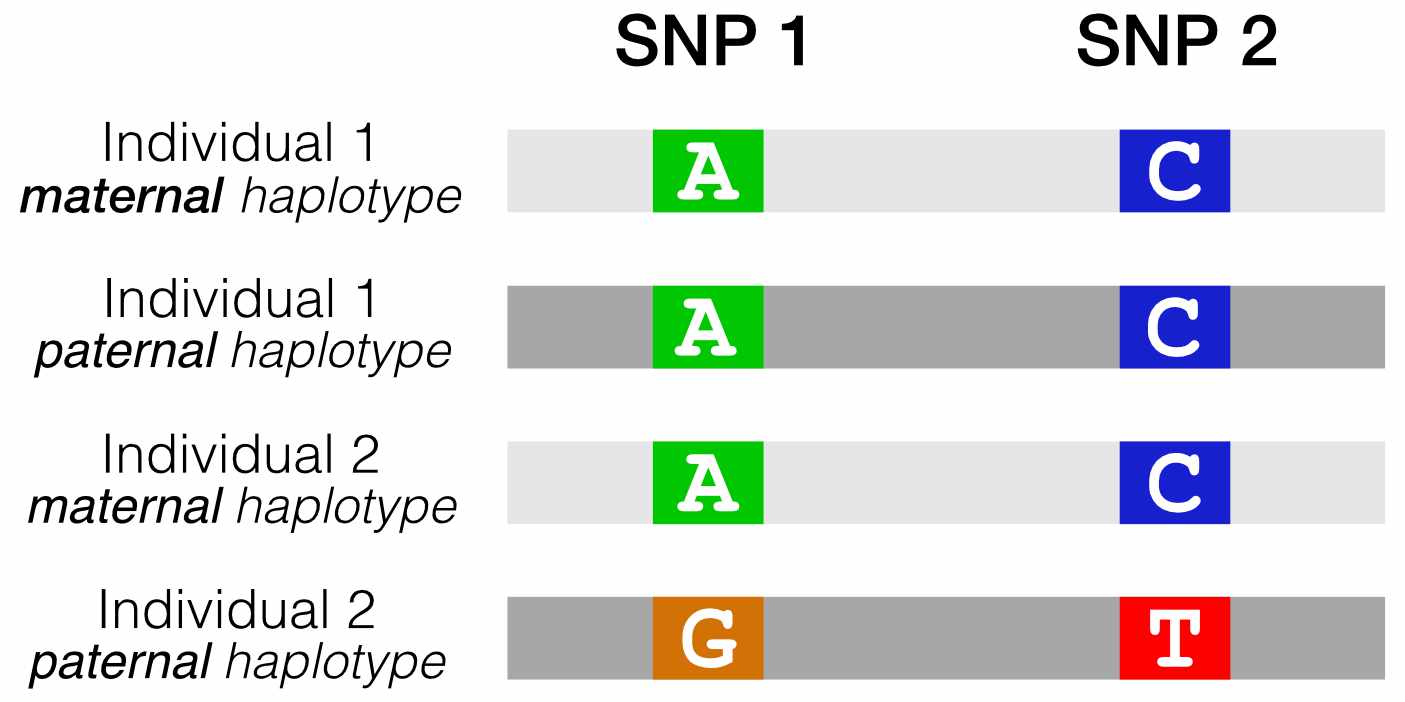
Fig. 3. Our reformatted VCF shows the combinations of alleles at two SNPs of interest, for all haplotypes in the 1000 Genomes dataset.