2.12 Interpreting IGV alignments
Sequencing reads in IGV are colored at bases where they differ from the reference genome. These differences can be caused by either real genetic variation or sequencing error. How would you distinguish these two?
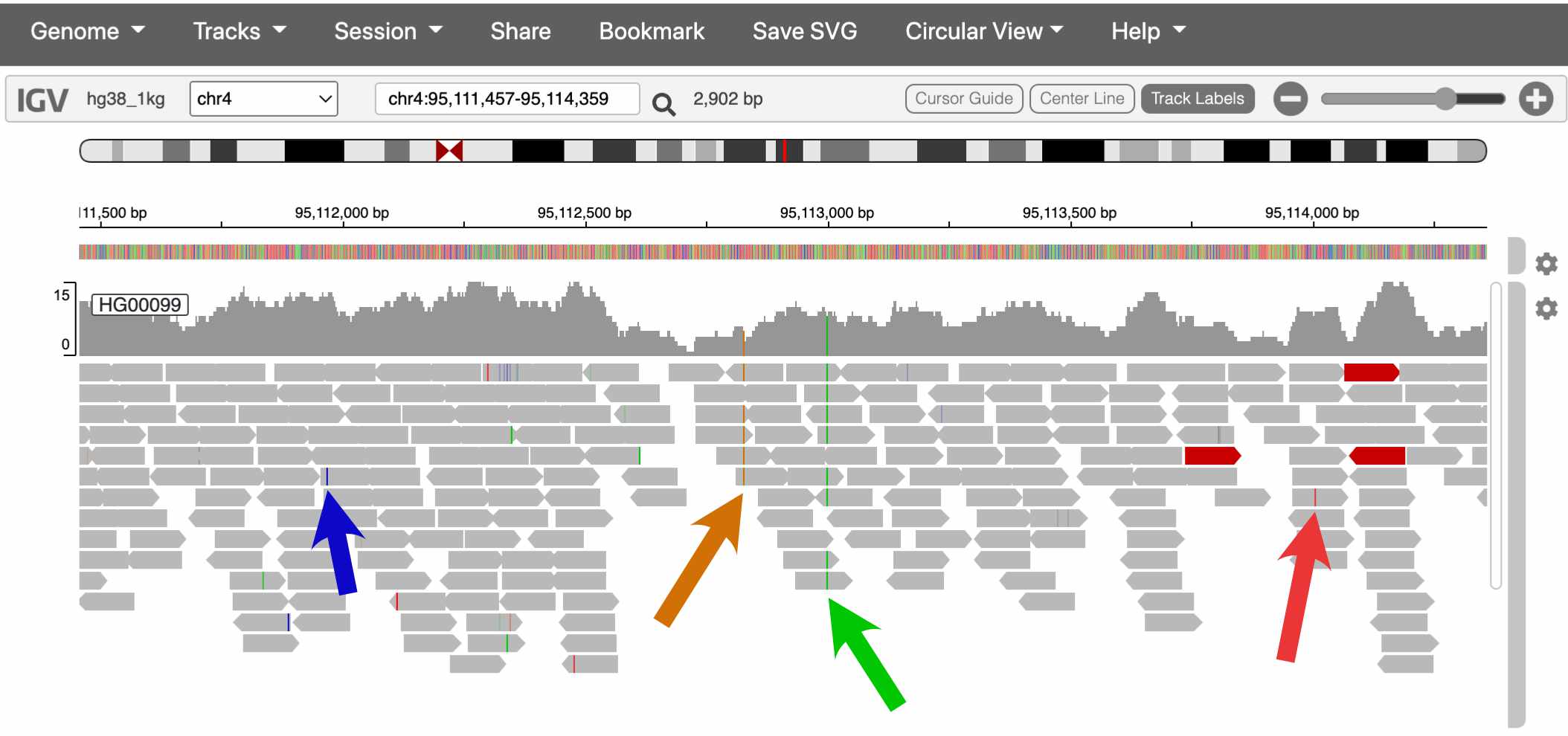
The sequencing coverage track also colors the positions that it thinks are real variants.
In the screenshot above, which spans about 2kb, there are two SNPsin the coverage track. This pattern holds more broadly through the genome – humans carry about one SNP every 1,000 bases.
Is one SNP every 1,000bp a lot or a little?
Humans actually have much lower amounts of genetic variation than many species, including many of the great apes.
This is mostly the result of human evolutionary history. Because the effective size of human populations has historically been low, with only very recent expansion, the gene pool is still fairly homogenous, with many rare variants and few common ones.