8.9 Phenotype data
Our phenotype for this GWAS is the \(\mathbf{IC_{50}}\) – the concentration of the GS451 drug that at which we observe 50% viability in cell culture.
## FID IID GS451_IC50
## 1 1001 1001 5.594256
## 2 1002 1002 8.525633
## 3 1003 1003 12.736739
## 4 1004 1004 12.175201
## 5 1005 1005 9.936742
## 6 1006 1006 9.163483The columns of this table are:
FID&IID: Family and individual IDs of the individualGS451_IC50: Measured \(\mathrm{IC_{50}}\) for the drug of interest
Plot the distribution of the phenotype.
## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.## Warning: Removed 1 row containing non-finite outside the scale range
## (`stat_bin()`).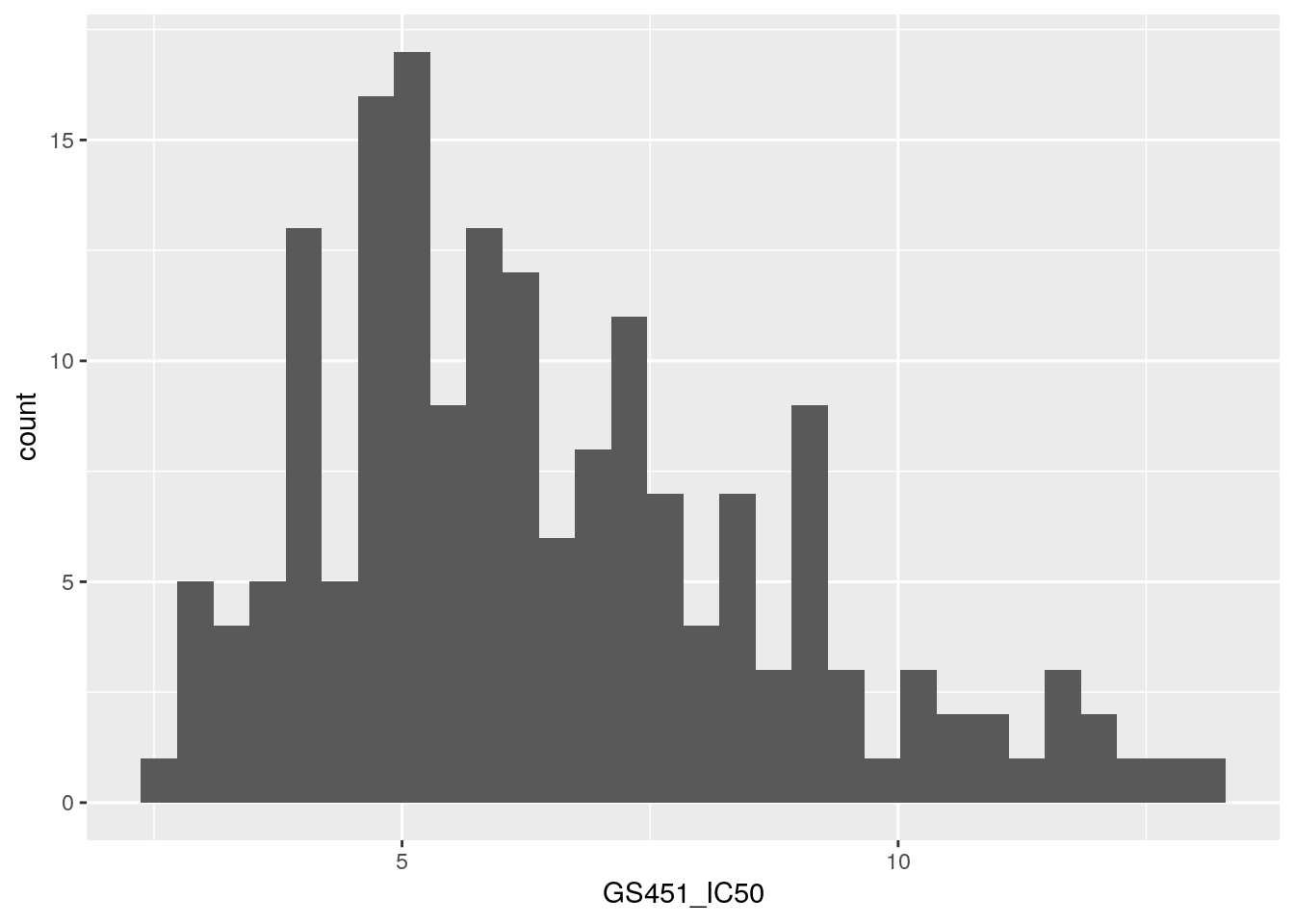
This data looks approximately normally distributed. This is important to check because this is one of the assumptions of linear regression, which we’ll be using to perform the GWAS.