2.5 Viewing one region of the genome
Once you hit enter, you should end up on a page like this:
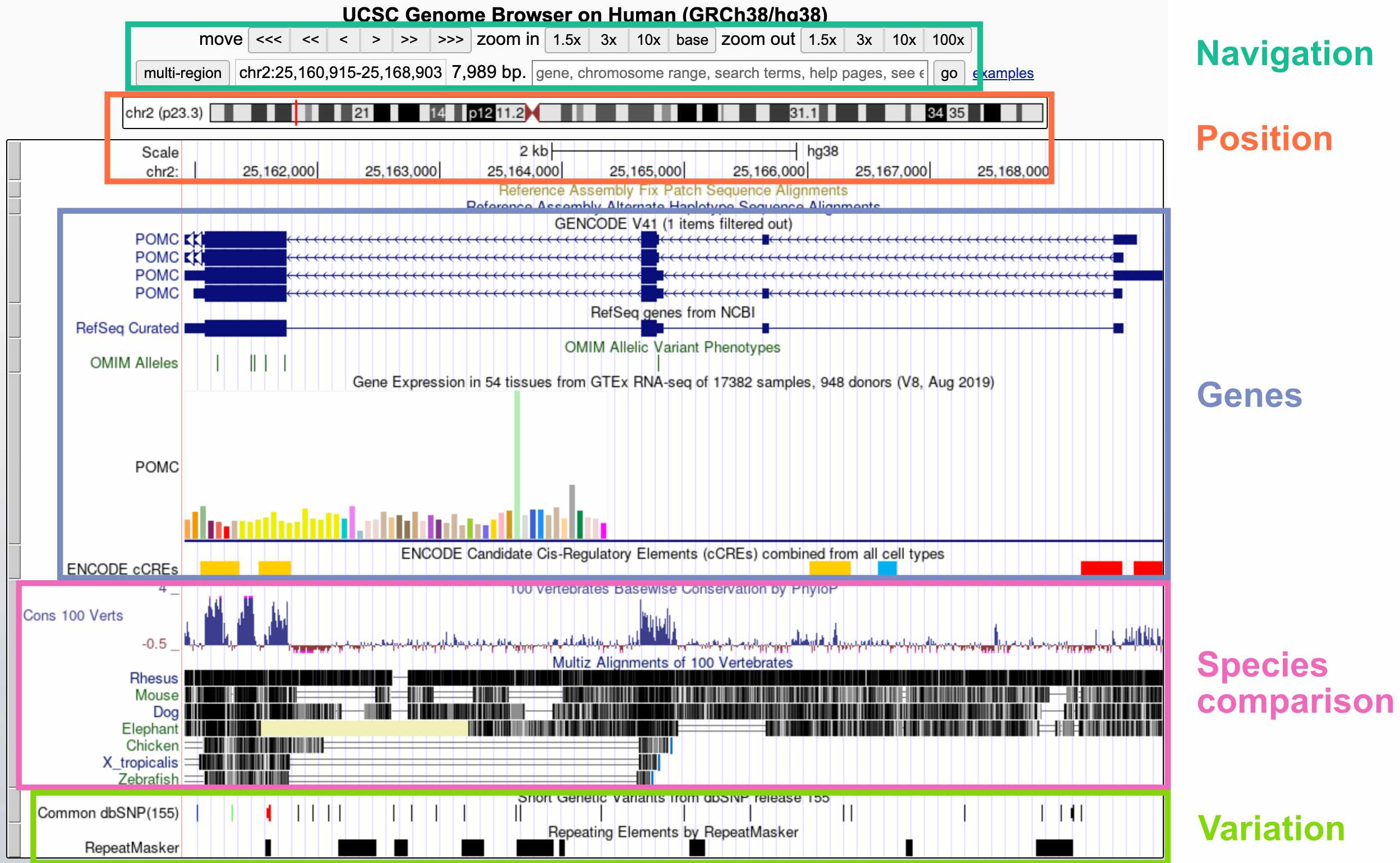
The default display includes these broad groups of annotations:
- Navigation: Buttons for zooming and moving around (you can also move by clicking the display, holding, and moving your mouse); current region; search bar
- Position: Current position on the chromosome; current base pair position
- Genes: Gene annotations; gene expression by tissue; gene regulatory elements (CREs)
- Species comparison: DNA sequence conservation across vertebrates; regions that align with the genomes of other vertebrates
- Variation: Genetic variants in the dbSNP database; repeat elements
Inspecting a specific track
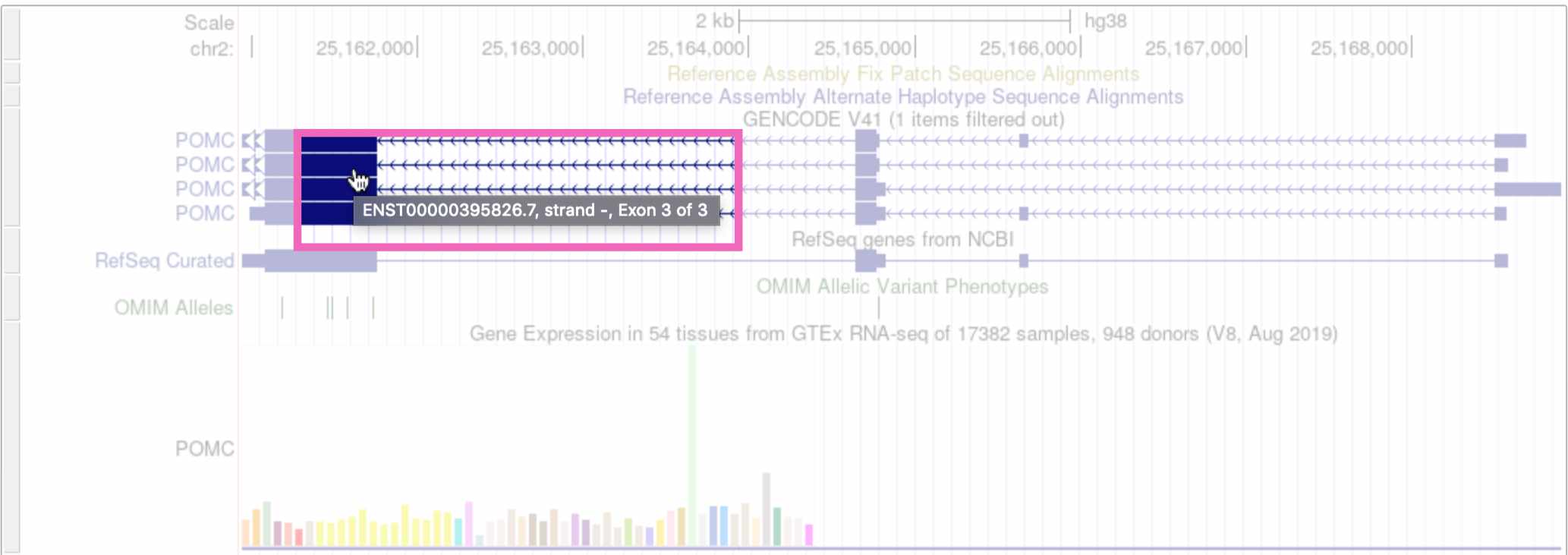
If you’re interested in more information about a specific track – for example, the POMC gene annotation – you can click on that element to go to a webpage with more details.
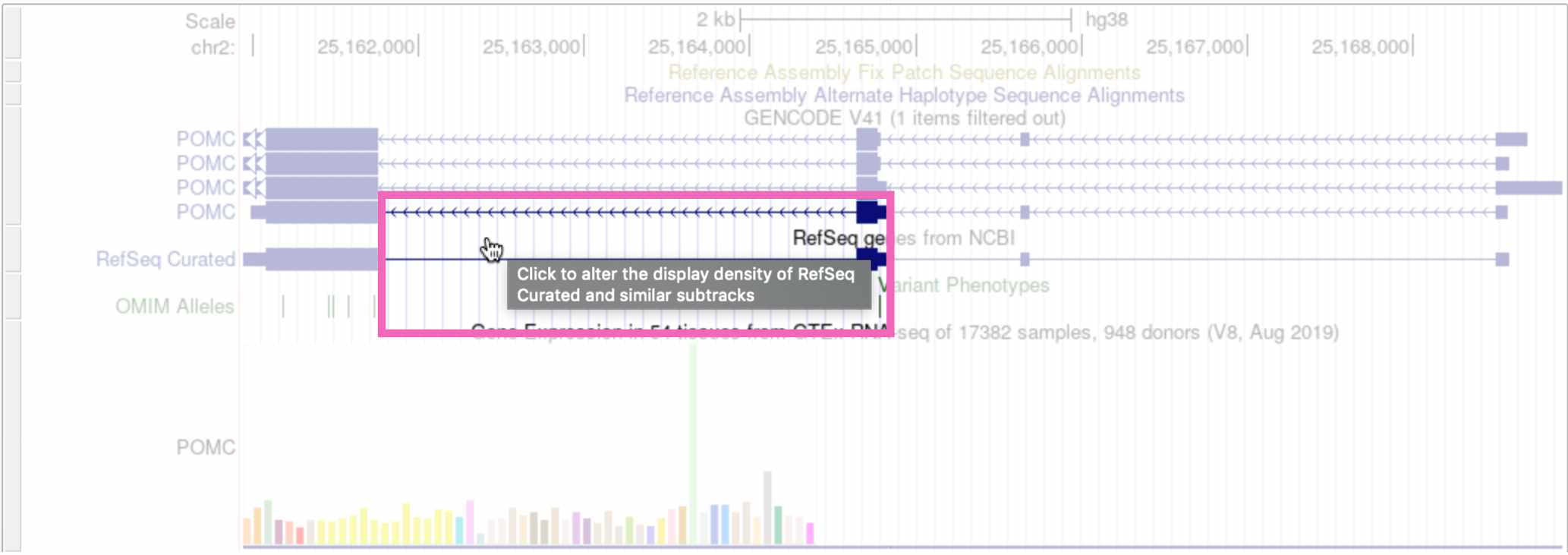
(Note that if you click on whitespace instead of an annotation element, it will change the track’s display density instead.)
Customizing the display tracks
The tracks that are automatically displayed are just a small subset of what’s available. You can select which tracks you want to see, and set their display density, by scrolling down on the page.
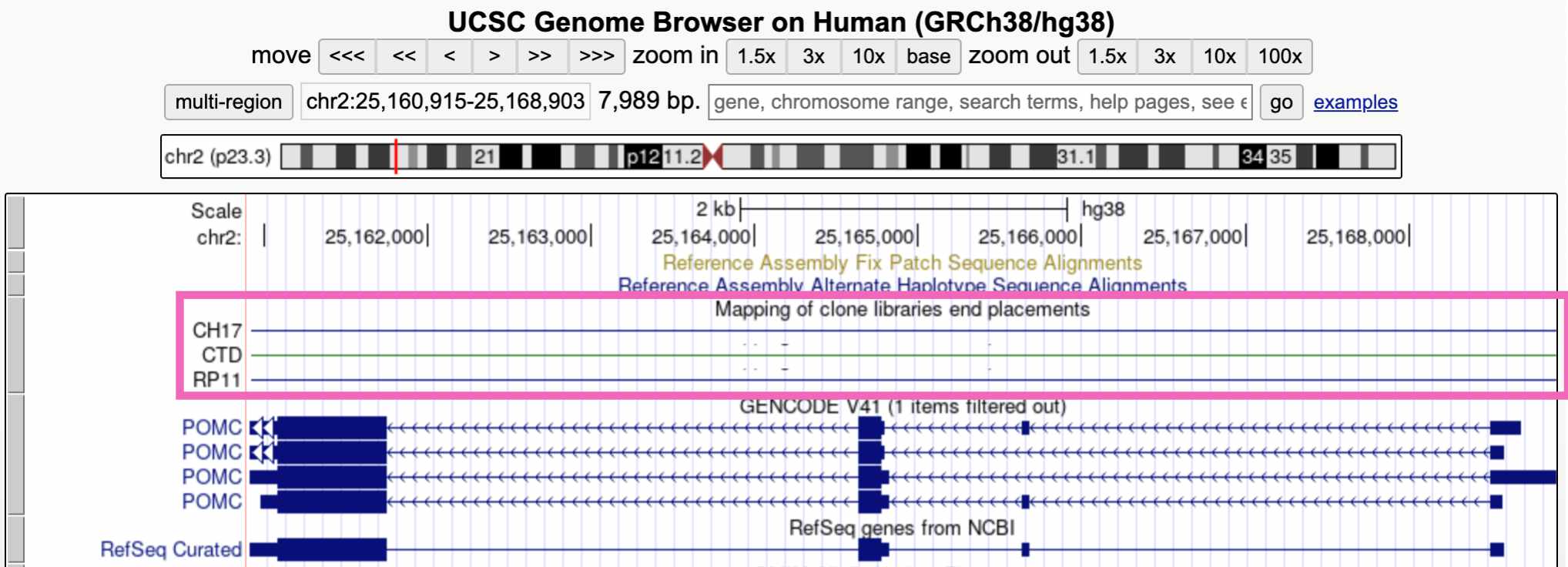
To add a new track to your browser view, click the drop-down menu below that track and select any of the options besides hide. Here we’re viewing the “Clone Ends” track, which shows the different individuals that were sequenced to create this section of the reference genome.
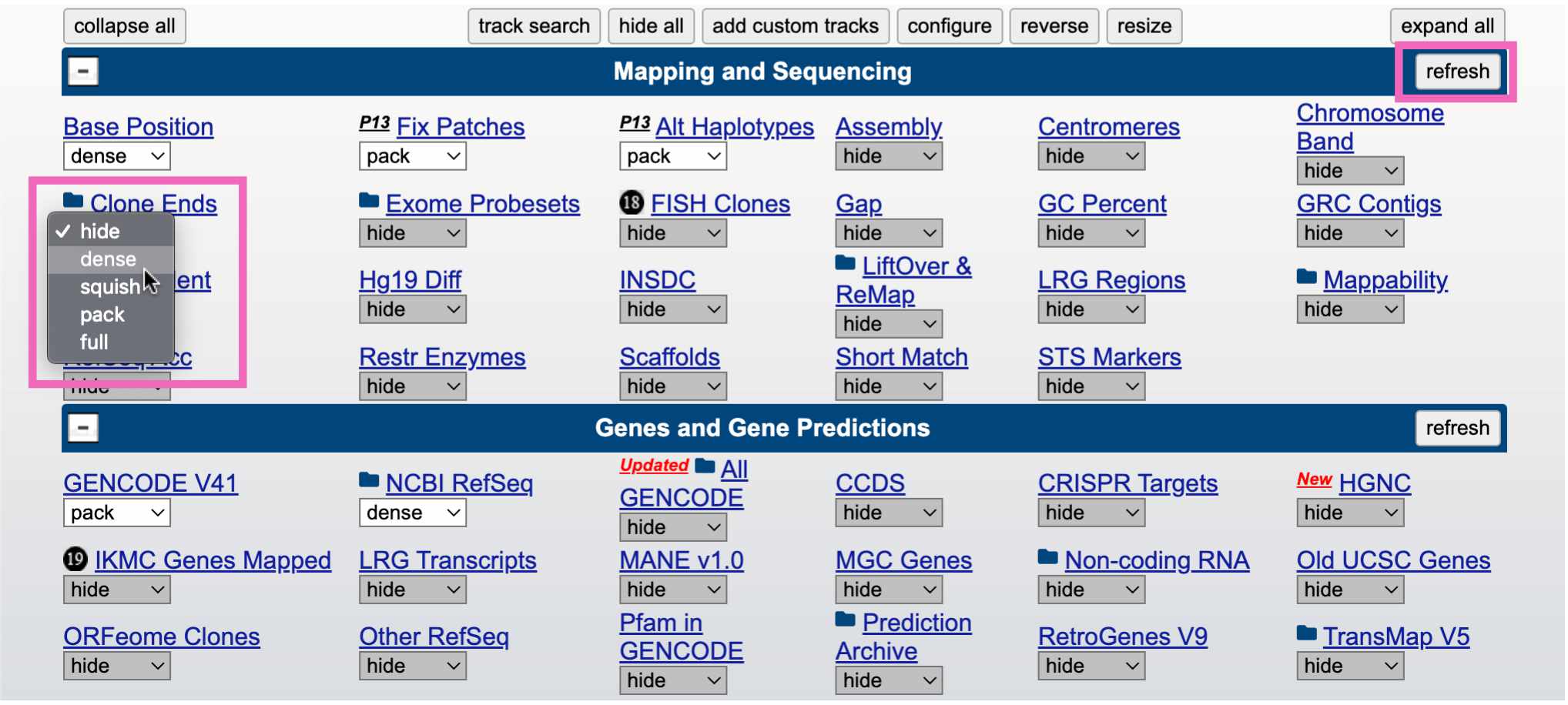
Click the refresh button in the upper right to reload the genome view. You should see something like this, showing that this region of the reference genome was sequenced in three individuals (CH17, CTD, and RP11):